Colorado State University
Module 6: Econ analysis in R
The purpose of this module is to cover some more advanced data manipulation techniques (combining datasets and reshaping data), show how to run basic regressions, and to provide an overview of other R capabilities that you may find useful in the future.
R has several built-in datasets for educational purposes, which we will use for the following exercises.
library(tidyverse)
library(stringr)
library(scales)
data() #show the build-in datasets in R
discoveries #Yearly Numbers of Important Discoveries (inventions/scientific), 1860-1959
LakeHuron #Water Level of Lake Huron, 1875-1972
lynx #Annual Canadian Lynx Trappings, 1821-1934
Since these data are stored as vectors, we first need to transform them into data frames.
discovery year <- data.frame(
year = 1860:1959,
num_disc = discoveries
)
head(discovery)
huron_levels <- data.frame(
year = 1875:1972,
huron_level = LakeHuron
)
head(huron_levels)
lynx_trappings<- data.frame(
year = 1821:1934,
num_trap = lynx
)
head(lynx_trappings)
1. Combining (Joining) Datasets
The dplyr package has a series of functions called joins (based on the SQL language). Join commands are used to combine rows from two or more datasets, based on a related column between them. This resource provides nice visual representations for how joins work.
1.1Inner Join
The most commonly used join is an inner_join(), which only keeps rows that match between two datasets (based on your matching variable). For the datasets we created above, each has the year variable that can be used to match observations. Since each of the datasets spans a different sequence of years, some observations will be dropped.
# Create a new dataset with discoveries and levels of Lake Huron by year
disc_huron <- inner_join(discovery, huron_levels, by = "year" )
head(disc_huron, 10)
If you want to join more than two datasets, you can tack on extra join statements with pipes (%>%).
# Join all three datasets
disc_huron_lynx <- inner_join(discovery, huron_levels, by = "year" ) %>%
inner_join(lynx_trappings, by = "year")
head(disc_huron_lynx, 10)
You can also join datasets using multiple matching variables. An example may look like this:
county_data <- inner_join(county_gdp, county_pop, by = c("year", "county"))
1.2 Left Join
A left_join() will return all rows from the left (first listed) dataset and the matched rows from the right dataset. R will automatically fill the “missing” (unmatched rows from the left) data with NA.
disc_huron_left <- left_join( discovery, huron_levels, by = "year")
head(disc_huron_left, 20)
1.3 Full Join
A full_join() is similar to the left_join, however all rows from both datasets are preserved. Again, the missing data is filled with NA.
disc_huron_full <- full_join( discovery, huron_levels, by = "year")
head(disc_huron_full, 10)
tail(disc_huron_full, 10)
1.4 Anti Join
To figure out why certain observations are being dropped, you can use anti_join(). An anti_join() will return only the observations from the left dataset that do not match any in the right dataset.
anti_join(huron_levels, discovery, by = "year")
2. Reshaping Datasets
Datasets are typically available in two different formats: wide and long. A wide format dataset contains values that do not repeat in the in the subject/identifier column. A long format dataset contains values that do repeat in the subject/identifier column.
Depending on your end goal, one format may be preferable to other. In general, the long format is better for storing data, running statistical models, and graphing in ggplot. Wide format data is easier to read and is generally better used for spreadsheets or tables in reports.
2.1 Pivot wider
The us_rent_income dataset has median yearly income and median monthly rate by state. It is stored in R in a long format, where estimate corresponds to the variable column and moe is a 90% margin of error. The pivot_wider() function transforms datasets from long format to wide format.
us_rent_income_wide <- us_rent_income %>%
pivot_wider(
names_from = variable,
values_from = c(estimate, moe)
)
head(us_rent_income_wide, 20)
2.2 Pivot longer
The relig_income dataset is in wide format, and it contains information on the number of individuals that fall within a certain income range based on their religion.
relig_income_long <- relig_income %>%
pivot_longer(
cols = -religion,
names_to = "income",
values_to = "count"
)
head(relig_income_long, 20)
3. Linear Regression
The lm() function is used to fit linear regression models. Regressions are typically saved as list objects that contain all the relevant information for model performance and inner workings.
Below is a simple example of how you would run a regression.
First, we retrive the dataset from R car package.
library(car)
help("Prestige")
View(Prestige)
In the simple linear regression example we have only one dependent variable(prestige) and one independent variable(education).
reg1 <- lm(prestige ~ education, data = Prestige)
summary(reg1) # print regression results
It's a useful practice to transform the variable to it's log form when doing the regression. You can either transform the variable beforehand or do so in the equation.
reg2 <- lm(prestige ~ education+log(income)+type, data = Prestige)
summary(reg2)
# save regression results as a data frame
regression_results <- data.frame(summary(reg2$coef)
regression_results <-round(regression_results, digit=3)
names(regression_results) <- c("coef", "stderr", "tval", "pval")
regression_results
You should always export the regression results as a table or figure, especially when you need to compare two or more different results. For lm(), the easiest package to use is stargazer.
library(stargazer)
stargazer(reg1, reg2, type = "text", title = "Regression Results",
align = TRUE, dep.var.labels = c("Prestige"),
covariate.labels = c("Education", "Log(Income)", "Type (Blue Collar)", "Type (Prof/Manager)"),
omit.stat = c("f", "ser"))
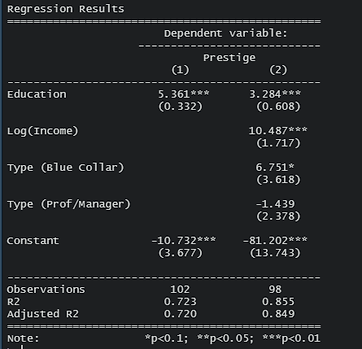
Personally, I prefer to generate a LaTeX table so I can render it directly in the output.
library(stargazer)
stargazer(reg1, reg2, type = "latex", title = "Regression Results",
align = TRUE, dep.var.labels = c("Prestige"),
covariate.labels = c("Education", "Log(Income)", "Type (Blue Collar)", "Type (Prof/Manager)"),
omit.stat = c("f", "ser"))
For more complicated regression models, I find the modelsummary package to be quite useful.
4. R Capabilities Beyond These Modules
Overall, these modules intended to provide a surface-level introduction to R and the functions that you will use most frequently as a graduate student in economics (if you choose to use R). Below are some examples of R’s other capabilities that may or may not be useful for you in the future.
4.1Statistical Modeling
R is first and foremost a programming language for statistical computing. R has many built-in regression models, however some very smart people have created some very useful packages for implementing complex statistical modeling techniques. Joey has used all of the following packages at some point in his research, and I have used most of them as well.:
The fixest package: Fast and user-friendly estimation of econometric models with multiple fixed-effects. Includes ordinary least squares (OLS), generalized linear models (GLM) and the negative binomial.
The ranger package: A fast implementation of Random Forests, particularly suited for high dimensional data. Ensembles of classification, regression, survival and probability prediction trees are supported.
The micEconAids package: Functions and tools for analyzing consumer demand with the Almost Ideal Demand System (AIDS) suggested by Deaton and Muellbauer (1980).
The cluster package: Methods for Cluster analysis. Much extended the original from Peter Rousseeuw, Anja Struyf and Mia Hubert, based on Kaufman and Rousseeuw (1990) “Finding Groups in Data”.
The MatchIt package: Selects matched samples of the original treated and control groups with similar covariate distributions – can be used to match exactly on covariates, to match on propensity scores, or perform a variety of other matching procedures.
4.2 Optimization
In mathematics, some equations cannot be solved analytically and require fancy numerical techniques to discover minimum and maximum values. This link provides an overview of all the different packages that provide solvers and numerical techniques for different optimization problems. As a caveat, these packages are not useful if you do not have a fundamental understanding of when they are necessary and the math behind the techniques.
In economics, these packages may be useful if you have a theoretical model that you cannot solve by hand. For example, they may be useful for simulating over different values of model parameters to see how the optima changes.
4.3 R Markdown
To conduct truly reproducible research, you need to be able to clearly communicate your process. Providing someone a folder filled with .csv and .R files can be useless unless the person fully understands your work flow.
All of these .html documents were created with R Markdown (poorly). Follow this link for an introduction for how to use R Markdown.
Also, read this.
4.4 Accessing Databases
RStudio can interact with local/remote databases in a manner similar to SQL using the odbc package. See this link for more information.
4.5 Accessing APIs
At some point you may be interested in using one of many publicly available data resources. In some cases, these data resources will have an application programming interface (API) that allows you to access/query data. This is a nice introduction to accessing API’s in R.
If you are lucky, someone has already created an easy to use package for accessing the API you are interested in. This is the case for NASS Quick Stats, a comprehensive database for agricultural data. The package rnassqs was built specifically for accessing NASS data in R. But you need to get your own API key from here.
4.6 Batch Downloading Data or Scraping Data
Unfortunately, not all publicly available data will have an API to query from. Oftentimes data is hosted on a website through several different file links or dashboards, and you want all of it. R can be used to batch download data from several links or to scrape data from websites. The following is an example from Joey.
"The most comprehensive county-level datasets for crop acreage in the state of California that I have found are the annual crop reports compiled by the California County Agricultural Commissioners. They are readily available by year in .pdf, .xls, and .csv formats. However, downloading all the files individually and combining them manually is both time consuming and bad for reproducibility. The following batch of code should download all the currently available data (as of right now, 2020 is the most recent available year). This should take at most 5 minutes to run (depending on internet connection), but it usually runs in about 30 seconds. It will combine all the data into one object called county_crop_data."
#These are the required packages
install.packages(c("RCurl", "XML"))
library(RCurl)
library(XML)
# First need to create a list of all .csv links on the webpage
url <- <- "https://www.nass.usda.gov/Statistics_by_State/California/Publications/AgComm/index.php"
html <- getURL(url)
doc <- htmlParse(html)
links <- xpathSApply(doc, "//a/@href")
free(doc)
csv_links<- links[str_detect(links, ".csv")]
# Clean up the links so the data can be easily downloaded
get_me <- paste("https://www.nass.usda.gov", csv_links, sep = "")
get_me <- unique(get_me)
# remove link for 2021, data is incomplete
get_me <- get_me[!grepl("AgComm/2021/", get_me)]
# Create a loop to import all the data one .csv at a time
crop_data <- list() # dump results here
for (i in get_me) {
temp <- read.csv(i)
names(temp) <- c( "year", "commodity", "cropname", "countycode", "county",
"acres", "yield", "production", "price_per", "unit",
"value")
crop_data[[length(crop_data)+1]] <- temp
}
# Append all the data and clean it
county_crop_data <- do.call(bind_rows, crop_data) %>%
# remove white space and correct misspelled county names
mutate(county = trimws(county),
county = ifelse(county == "San Luis Obisp", "San Luis Obispo", county)) %>%
# for now remove all years prior to 1989, remove state totals, and remove missing data
filter(str_detect(county, "State|Sum of") == FALSE,
is.na(acres) == FALSE)
head(county_crop_data)
This example isn’t really web scraping, because all of the data is available in readily downloadable tables. R is certainly capable of web scraping, but Python is a more commonly used language for scraping.
4.6 GIS
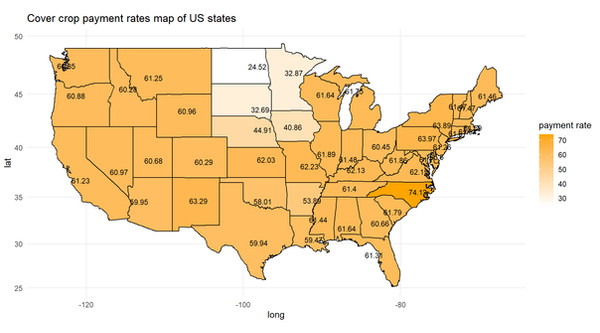
R is capable of handling spatial analysis and visualization. This book is a good resource. Most anything that you can accomplish with ArcGIS or QGIS can also be accomplished in R. Here is a simple example of how I drew the cover crop payment rates in EQIP by state in the U.S. in 2023.
library(readxl)
#Load the data I need to draw the map
ccmap<-read_xlsx("C:/Users/wangm/OneDrive - Colostate/dissertation/CH1/2023CH1/Data/cover crop payment.xlsx")
#input the data that matching the state fips codes with state names
state_fips<- read_xlsx("C:/Users/wangm/OneDrive - Colostate/dissertation/CH1/2023CH1/Data/state_Region.xlsx")
state_fips<- subset(state_fips, select = c("state","Name")) # clean the data, leave the columns that I need
colnames(ccmap)[1]<- "Name" #Rename the column to match the column name in state_fips
ccmap<-merge(ccmap,state_fips,by="Name") # merge the data, this is inner joint
ccmapccmap<-subset(ccmap,select = c("state","cc"))
# Now I have a clean dataset, with state fips code, and the covercrop payment rate
library(maps) # Built in geo data in R
states_map<- map_data("state") # But there is only state names in the geo dataset
state_fips$Name<-tolower(state_fips$Name) #all letters to lowercase
states_map<-merge(states_map,cstate_fips, by.x = "region", by.y = "Name")
states_map<-merge(states_map,ccmap,by="state")
library(dplyr)
states_map <- arrange(states_map, group, order)
colnames(states_map)[8]<-"value"
state_centroids <- states_map%>%
group_by(region) %>%
summarise(long = mean(long), lat = mean(lat), value = mean(value))
ggplot() +
geom_polygon(data = states_map, aes(x = long, y = lat, group = group, fill = value), color = "black") +
geom_text(data = state_centroids, aes(x = long, y = lat, label = round(value, 2)), size = 3, color = "black") +
coord_map() +
scale_fill_gradient(low = "white", high = "orange") +
theme_minimal() +
labs(fill = "payment rate",
title = "Cover crop payment rates map of US states")